Just as I was sitting down, deciding what to write this post on, someone asked me on twitter "What are SNPs"?
Every profession and area of study has a bunch of acronyms and field specific terms that can be very confusing to those outside the field. Genetics is no different and SNP is one of our worst.
SNP stands for Single Nucletide Polymorphism.
No clearer, huh?
Okay, lets start at the end. Polymorphisms are when there is more that one version of a gene in the population - think eye colour and have a look at my post here.
When we talk about Single Nucleotide Polymophisms it means that at one specific position in the genome, some individuals will have one nucleotide and some individuals will have a different nucleotide.
There are a huge number of positions where this occurs and they're all categorised in a database. Many of the differences have no apparent function, others cause changes but neither option is better than the others. Other rare SNPs may be involved in genetic diseases.
SNPs are very useful in genetics for a number of reasons. One of the big uses in is Genome Wide Association Studies, which allow us to find regions of the genome which are associated with certain traits or diseases. The SNPs themselves may not actually be involved, but nearby regions maybe. We'll talk more about this when I get onto recombination.
Another reason why it's important to know about SNPs is in looking for the genetic mutation causing someone's disease. When their gene is sequenced and compared to the human reference genome, a number of different mutations may come up. It's important to be able to rule out the ones that we know are common and are therefore unlikely to be causing the disease.
So that's it, SNPs demystified. They're just single nucleotide substitutions, we just gave them a complicated name.
A project to explain topics in genetics to people who don't have any formal training in it.
Monday, February 24, 2014
Monday, February 17, 2014
Convergent Evolution
I've been working on a project recently which involves convergent evolution, so I thought I'd talk about that.
When we see the same trait in different species it is often because these species have an ancestor in common that developed the trait, passing it on to the species that later develop from it. This is similar to a parent passing on a trait to their children, such as eye colour.
A nice trait to think of in this way is the ability to incubate young in the womb versus laying eggs.
We assume that egg laying was the original condition and that an ancestor species of mammals (another term for mammals which incubate their young in-utero is Eutherians) developed the ability to incubate young in its womb instead - a feature which has a lot of benefits for the safety of the unborn young, such as protection from the elements or from predators. From phylogenetic studies we know that all mammals have a common ancestor which they don't share with reptiles, birds or fish - all of which lay eggs.
There are other species which help to show the development of this trait - Marsupials, for example, which don't lay eggs but incubate their young in external pouches, such as the kangaroo. There is also a further group of mammals which lay eggs (Monotremes), the platypus is an example of this. They are grouped with other mammals because of other mammalian traits they have, such as warm bloodedness, but they show us an intermediate stage of the development of the ability to incubate their young in the womb.

(image from http://palaeo.gly.bris.ac.uk/palaeofiles/marsupials/Index.htm)
Anyway, that was a bit of a sidetrack, because that trait is not an example of convergent evolution. Convergent evolution is when, rather than multiple species inheriting a trait from an ancestor species, the trait evolves independently in different species.
An example of this is the ability to fly. Both birds and bats possess the ability to fly. However, the bat is a mammal and much more closely related to other, non-flying mammals than it is to bats. If you only know this information it would be possible to assume that flight was lost in other mammals and it was an ancestral trait retained in bats. However, if you look at the structure of a bird and a bat wing, you can see that they are very different.

(image from http://evolution.berkeley.edu/evosite/evo101/IIC1Homologies.shtml)
With some traits, like flight, it is fairly simple to determine if covergent evolution has occurred. However with other traits it can be much more difficult, at which point a comparison at the DNA level is needed to determine the similarity between species.
When we see the same trait in different species it is often because these species have an ancestor in common that developed the trait, passing it on to the species that later develop from it. This is similar to a parent passing on a trait to their children, such as eye colour.
A nice trait to think of in this way is the ability to incubate young in the womb versus laying eggs.
We assume that egg laying was the original condition and that an ancestor species of mammals (another term for mammals which incubate their young in-utero is Eutherians) developed the ability to incubate young in its womb instead - a feature which has a lot of benefits for the safety of the unborn young, such as protection from the elements or from predators. From phylogenetic studies we know that all mammals have a common ancestor which they don't share with reptiles, birds or fish - all of which lay eggs.
There are other species which help to show the development of this trait - Marsupials, for example, which don't lay eggs but incubate their young in external pouches, such as the kangaroo. There is also a further group of mammals which lay eggs (Monotremes), the platypus is an example of this. They are grouped with other mammals because of other mammalian traits they have, such as warm bloodedness, but they show us an intermediate stage of the development of the ability to incubate their young in the womb.
(image from http://palaeo.gly.bris.ac.uk/palaeofiles/marsupials/Index.htm)
Anyway, that was a bit of a sidetrack, because that trait is not an example of convergent evolution. Convergent evolution is when, rather than multiple species inheriting a trait from an ancestor species, the trait evolves independently in different species.
An example of this is the ability to fly. Both birds and bats possess the ability to fly. However, the bat is a mammal and much more closely related to other, non-flying mammals than it is to bats. If you only know this information it would be possible to assume that flight was lost in other mammals and it was an ancestral trait retained in bats. However, if you look at the structure of a bird and a bat wing, you can see that they are very different.
(image from http://evolution.berkeley.edu/evosite/evo101/IIC1Homologies.shtml)
With some traits, like flight, it is fairly simple to determine if covergent evolution has occurred. However with other traits it can be much more difficult, at which point a comparison at the DNA level is needed to determine the similarity between species.
Saturday, February 8, 2014
Mendelian genetics - that bit with the monk
Gregor Mendel was an Augustinian monk who lived in the 19th Century. He gave us some of the first quantifiable evidence for genetic inheritance and the type of genetics he described is named after him.



Mendel was a gardener at the monastary, he grew peas. While he was growing the peas, he observed what happened when you breed a plant with one trait against a plant with a different trait. What he found was a set of ratios that are always the same in the offspring.
This is the bit where we could come to those crosses you might remember from high school, but I'm actually going to leave that for another day. I will, however try to explain a bit about Mendelian genetics without resorting to drawing crosses.
Every gene that you have has two copies (unless your a guy, then you X & Y makes thing a bit more complicated). This is because you have paired chromosomes like this:

One of your copies came from your mother and one from your father and they can be slightly different (the different copies are called alleles). For example your parents may have had different coloured eyes, so they might be passing you slightly different version of the eye colour genes. Don't forget that your parents had two copies as well - so they might have different types which could be passed to you.
Let's say you received a blue eye colour allele from your father and a brown eye colour allele from your mother. How can we predict what colour eyes you have?
Well, Mendelian genetics relies on the concept of recessive and dominant alleles. A dominant allele only has to have one copy to show, but a recessive allele need two copies to show. In the case of your eye colour, we know from experiments that the brown allele is dominant and the blue allele is recessive. This means that you would have brown eyes.
Mendel noticed that certain traits in his peas were dominant and some were recessive. Traits he looked at included things like: shape (wrinkled or round) and seed colour (yellow or green).
Unfortunately, although Mendel published his work at the time, it didn't reach the wider scientific community and had to wait until the early 20th century when the same conclusions were independently discovered by other researchers. Previously to this work being discovered it was assumed that traits in the offspring were a merging of the traits in the parents rather than a case of either/or.
The other interesting point to make is that, knowing what we now know about statistics and probability, it appears that Mendel may have either tweaked some of his results or played around with the plants until he got ones which showed exactly the results he wanted - the ratios are just too good, it wouldn't happen in a modern experiment. However, we shouldn't really judge him too harshly, biology in the 19th century was mostly observation and very little planning of experiments, so he probably wasn't doing anything that the others wouldn't have been doing.
Monday, February 3, 2014
Phylogeny or the Tree of Life
The tree of life was originally suggested by Darwin as a way of explaining the relationships between different species. It's a lot like a family tree in both form and function. The study of these trees is called phylogenetics.
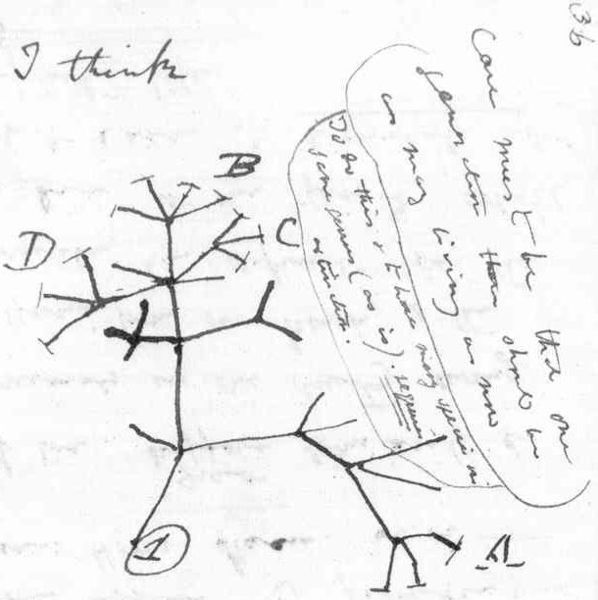
A modern phylogenetic tree (that's the correct term) looks something like this:

If we look at the group at the bottom of the tree, these are all apes (humans count in this group too). We can see that Bonobos and Chimpanzees are the most closely related, with Humans next to them.
One of the important things that people often get wrong about evolution is the idea that we 'descended from monkeys'. If you look at the tree, each of the tips represents a modern species, but the next branch up, from where two species combine, represents a species that was somewhat similar to each of the modern species, but it wasn't exactly the same as any of them. Sometimes we have fossil records for these species, sometimes we don't. So when people say 'we descended from monkeys', it is closer to the truth to say both monkeys and humans descended from a common ancestor species in the fairly recent past (on an evolutionary timescale).
If you're interested in learning more about 'common ancestor' species. There's a fun website called Timetree.org which will show you the time at which researchers think that the speciation event (the branching that you can see on the tree) happened for different pairs of species. This gives you an idea of when the common ancestor species might have lived.
You can also find out some interesting facts about which animals are most closely related to each other. Some that I find amusing are:
Dogs vs Cats - 55 Million years ago
but
Dogs vs Pandas - 45 Million years ago (you'll have to use the latin name for Panda - Ailuropoda)
Cows vs Horses - 82 Million years ago
but
Cows vs Whales - 56 Million years ago

A modern phylogenetic tree (that's the correct term) looks something like this:

If we look at the group at the bottom of the tree, these are all apes (humans count in this group too). We can see that Bonobos and Chimpanzees are the most closely related, with Humans next to them.
One of the important things that people often get wrong about evolution is the idea that we 'descended from monkeys'. If you look at the tree, each of the tips represents a modern species, but the next branch up, from where two species combine, represents a species that was somewhat similar to each of the modern species, but it wasn't exactly the same as any of them. Sometimes we have fossil records for these species, sometimes we don't. So when people say 'we descended from monkeys', it is closer to the truth to say both monkeys and humans descended from a common ancestor species in the fairly recent past (on an evolutionary timescale).
If you're interested in learning more about 'common ancestor' species. There's a fun website called Timetree.org which will show you the time at which researchers think that the speciation event (the branching that you can see on the tree) happened for different pairs of species. This gives you an idea of when the common ancestor species might have lived.
You can also find out some interesting facts about which animals are most closely related to each other. Some that I find amusing are:
Dogs vs Cats - 55 Million years ago
but
Dogs vs Pandas - 45 Million years ago (you'll have to use the latin name for Panda - Ailuropoda)
Cows vs Horses - 82 Million years ago
but
Cows vs Whales - 56 Million years ago
Subscribe to:
Posts (Atom)